Fast custom image filters using low level callables

scipy’s generic_filter
is pretty neat. Give it a function and a filter footprint, and it will apply
the function at every footprint in the image. The trouble is that it can
be a bit slow:
In [1]: from scipy.ndimage import generic_filter
In [2]: from skimage import data
In [3]: from skimage.morphology import disk
In [4]: image = data.camera()
In [5]: timeit generic_filter(image, max, footprint=disk(11))
14.3 s ± 134 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)Yikes. But how about numpy.max instead?
In [6]: import numpy as np
In [7]: timeit generic_filter(image, np.max, footprint=disk(11))
1.88 s ± 13.7 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)Better, but still not great, especially when compared against the existing maximum filter:
In [8]: from scipy.ndimage import maximum_filter
In [9]: timeit maximum_filter(image, footprint=disk(11))
218 ms ± 4.37 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)I want to use this simple maximum filter example to show to build a low
level callback
(i.e., a lightly wrapped compiled C function) to be used with generic_filter
in order to get better performance from custom filters (beyond just max).
The scipy docs
provide a number of examples using geometric_transform to illustrate
low level callbacks. I highly recommend reading those.
Building the low level callback
If we read the Notes section in the docstring for generic_filter we see:
This function also accepts low-level callback functions with one of the following signatures and wrapped in scipy.LowLevelCallable:
int callback(double *buffer, npy_intp filter_size, double *return_value, void *user_data) int callback(double *buffer, intptr_t filter_size, double *return_value, void *user_data)
The docs go on to provide more details, but here is the summary: we’ll write a
C function having the given signature that transforms buffer (the array of
image values currently under the filter footprint) to produce a return value.
Let’s write some C.
FILE: nice_filters.c
#include <math.h>
#include <stdint.h>
int max_filter(
double * buffer,
intptr_t filter_size,
double * return_value,
void * user_data
) {
double max = -INFINITY;
for(int i=0; i<filter_size; i++) {
if (buffer[i] > max) {
max = buffer[i];
}
}
*return_value = max;
// return 1 to indicate success (CPython convention)
return 1;
}Then compile it to a shared library:
gcc -shared -fpic nice_filters.c -o nice_filters.soOK. Now let’s load it with ctypes and wrap it as a low level callable:
In [10]: import ctypes
In [11]: from scipy import LowLevelCallable
In [12]: clib = ctypes.cdll.LoadLibrary('./nice_filters.so')
In [13]: clib.max_filter.restype = ctypes.c_int
In [14]: clib.max_filter.argtypes = (
...: ctypes.POINTER(ctypes.c_double),
...: ctypes.c_long,
...: ctypes.POINTER(ctypes.c_double),
...: ctypes.c_void_p,
...: )
In [15]: max_filter_llc = LowLevelCallable(clib.max_filter)Now max_filter_llc can be used in generic_filter. Let’s time it!
In [16]: timeit generic_filter(image, max_filter_llc, footprint=disk(11))
422 ms ± 7.56 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)Not bad. Although given that scipy’s maximum_filter still runs in half the
time, there’s apparently some overhead to the generic_filter framework.
Does it work?
In [18]: f1 = maximum_filter(image, footprint=disk(11))
In [19]: f2 = generic_filter(image, max_filter_llc, footprint=disk(11))
In [20]: abs(f1-f2).max()
Out[20]: 0Cool.
Note that we haven’t talked about the user_data parameter. This parameter
allows for adding extra arguments (e.g., a global threshold value) to be used
by the callback. The scipy docs
provide some example usage that’s worth checking out.
Although, not all extra arguments can be plugged in as user-provided data. For example, as far as I can tell, it’s not possible to get the current position of the filter as an argument to the callback, which would be nice to optionally have in order to create position dependent filters. Maybe I will dive into the ndimage source to see if this sort of extension is feasible when I have some free time.
Bonus: sliding window standard deviation filter
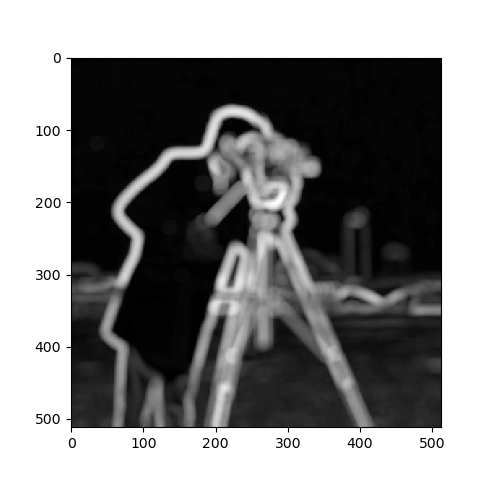
We went through all the rigmarole, so let’s at least write a filter that might
not be readily available in pre-written routines. The following computes the
standard deviation of the values under the filter footprint and uses that as
the filter response. So, using this with generic_filter computes a sliding
window standard deviation filter. This produces something not unlike a gradient
magnitude image, i.e., it produces large (bright) values where the image
changes intensity and small (dark) values where the image is homogeneous.
int std_filter(
double * buffer,
intptr_t filter_size,
double * return_value,
void * user_data
) {
double mu = 0;
double std = 0;
// Compute the empirical mean under the footprint
for(int i=0; i<filter_size; i++)
mu += buffer[i] / filter_size;
/// Compute the empirical standard deviation under the footprint
for(int i=0; i<filter_size; i++)
std += (buffer[i] - mu)*(buffer[i] - mu) / (filter_size-1);
*return_value = sqrt(std);
return 1;
}