Calling Fortran from Python with f2py
A short guide on using f2py to call Fortran subroutines from Python
Preliminaries
It is assumed that you have Python, NumPy, and Matplotlib installed. If not, you can find many instructions around the web for installing these.
Python is a scripting language, while Numpy and Matplotlib are Python modules (or libraries) for scientific computing and plotting respectively. This is not a guide specificly on using these libraries — although, they are great, and you should try them out in more detail.
f2py is a command line utility that is included with Numpy that converts files containing Fortran subroutines or modules into Python modules. This allows you to code your numerical routines in Fortran, while allowing Python scripts to “drive” the main program for plotting, etc. You can find more detail about f2py by reading its User guide and reference manual.
Reading text image data from a file and thresholding it in Fortran
As an example, we will
- Read a square image stored in a file as text data into Python as a double precision matrix.
- Pass the image to a Fortran subroutine that thresholds the values of the matrix.
- Return the image back to the Python script and plot the results.
Writing the Fortran routine
Let’s assume we have a file, my_lib.f90, containing one or more Fortran subroutines that looks like the following:
subroutine threshold_image(image, n, threshold, output)
! Inputs: image, n, threshold.
! Output: output
! output(i,j) is 1 if image(i,j) > threshold and 0 otherwise.
integer n
real(8) threshold
real(8), dimension(n,n) :: image, output
!f2py intent(in) :: image, threshold
!f2py intent(hide), depend(image) :: n = shape(image, 0)
!f2py intent(out) output
write(*,*) "Hello from the Fortran subroutine!"
! Loop through columns and rows and threshold the image.
do j=1,n
do i=1,n
if (image(i,j) > threshold) then
output(i,j) = 1.0
else
output(i,j) = 0.0
end if
end do
end do
end subroutineThis subroutine has 3 special comments that start with !f2py:
- The first tells
f2pythat the variablesimageandthresholdare required inputs when called from Python. - The second tells
f2pythat the variablenis defined implicitly through theimageargument when called from Python, and its value is the size of the first dimension ofimage - The third tells
f2pythat the variable,output, does not need to be provided as an argument when called from Python, and the variable is in fact returned by the function when called from Python. If multiple variables are returned, they are returned as a Python Tuple.
Compiling the Fortran file to a Python module
From the command line where the file is present, run
f2py -c -m my_lib my_lib.f90A new file is produced, my_lib.so, which can be imported into Python.
Testing the routine from a Python script
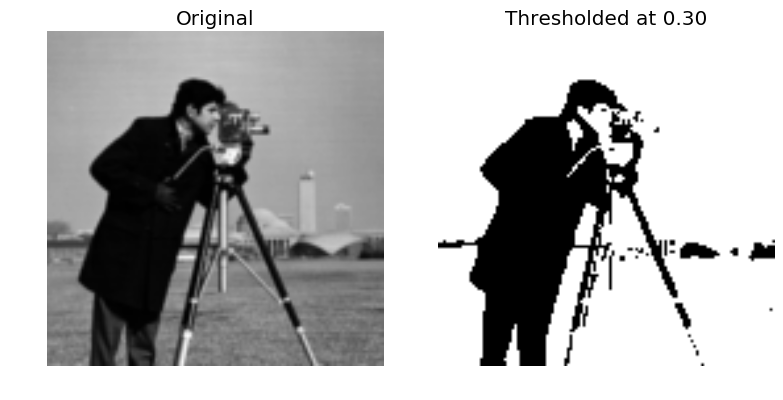
Let’s take a square image stored as a text file, and test our routine. Write a file, main.py, containing:
import numpy as np
import matplotlib.pyplot as plt
import my_lib as ml
# Read matrix from text file as double precision matrix.
I = np.genfromtxt('./image.txt', dtype=np.float64)
# Threshold value.
t = 0.3
# Call the fortran routine.
T = ml.threshold_image(image=I, threshold=t)
# Plot the images.
fig, axes = plt.subplots(1, 2)
axes[0].imshow(I, cmap=plt.cm.gray)
axes[0].axis('off'); axes[0].set_title('Original')
axes[1].imshow(T, cmap=plt.cm.gray)
axes[1].axis('off'); axes[1].set_title('Thresholded at %.2f' % t)
plt.tight_layout()
plt.show()And run it from the command line:
python main.pyIf successful, you should see both the original and thresholded image displayed as well as a the line “Hello from the Fortran subroutine!” printed.